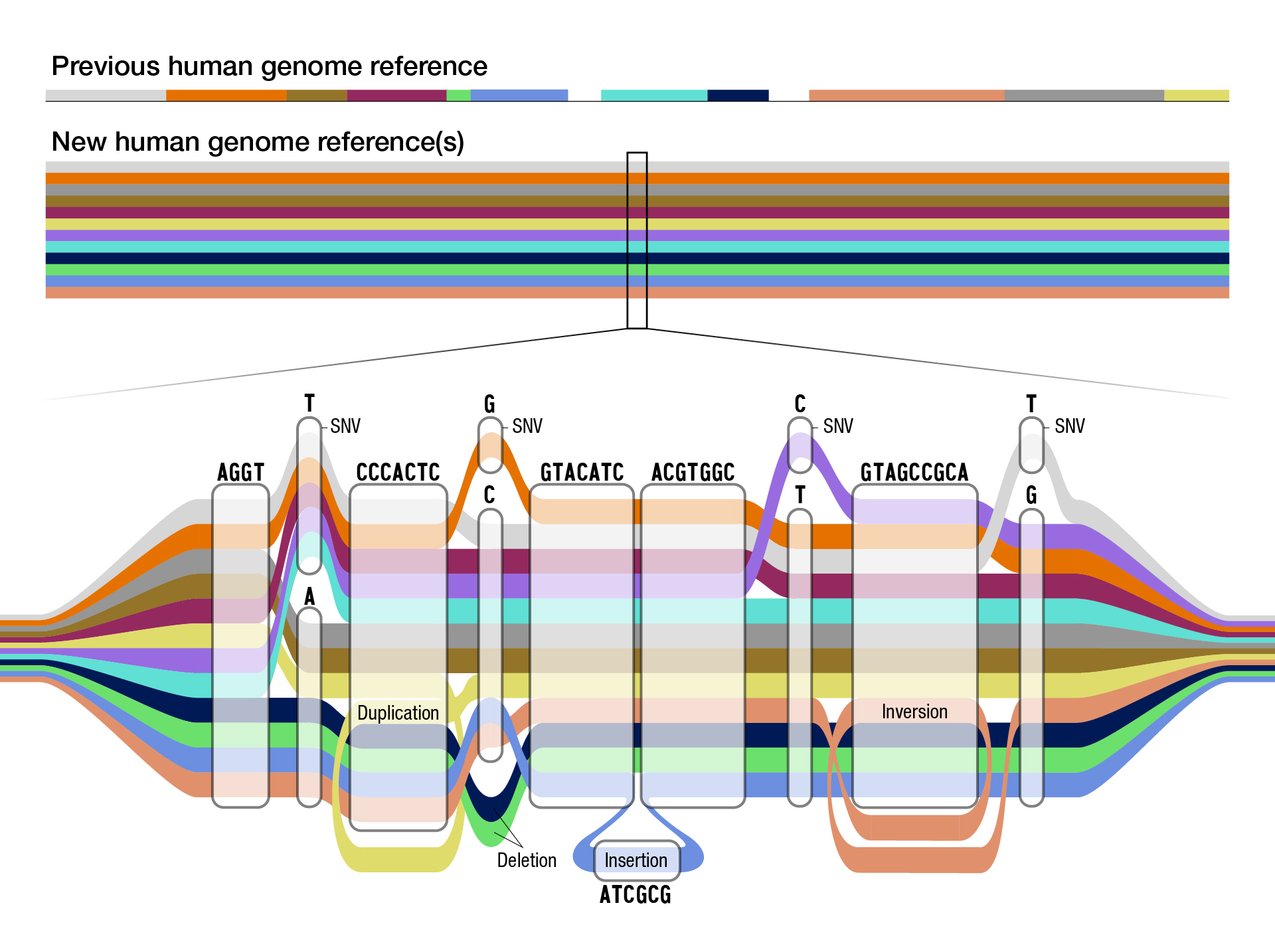
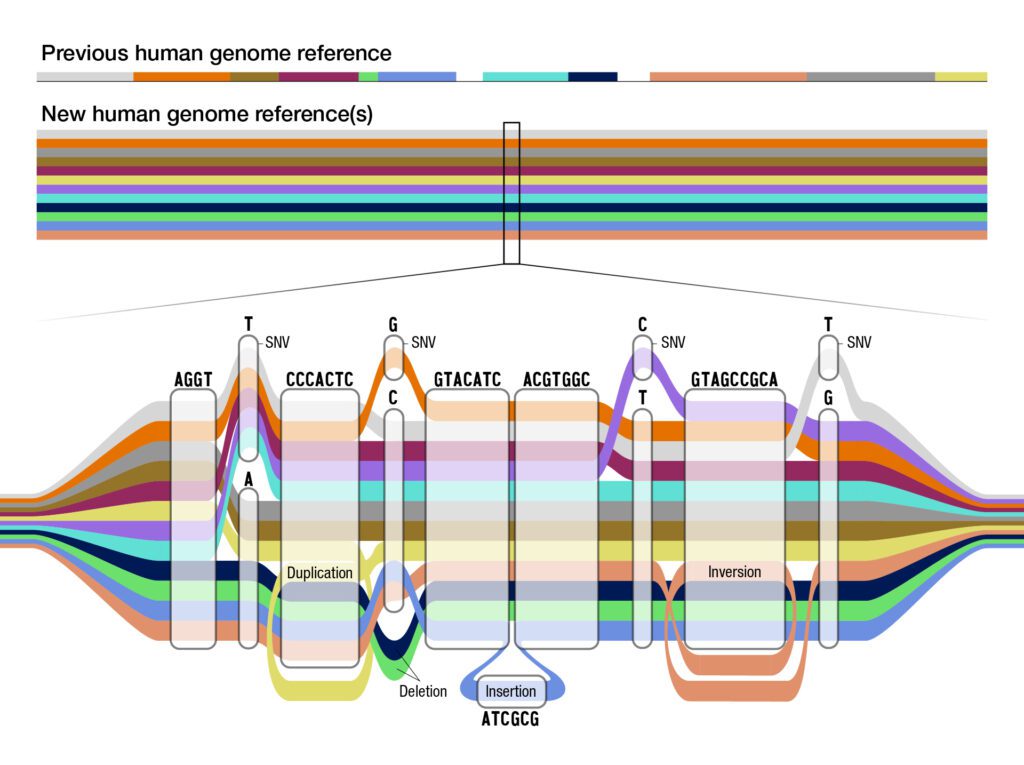
The National Human Genome Research Institute (NHGRI) released the “pangenome,” the updated map of the human genome, which includes more diverse DNA than ever before. This development could have important implications for biopharmaceutical research and specifically addressing disparities in medicine.
The updated “pangenome” includes DNA sequencing based on 47 individuals. “With each person carrying a paired set of chromosomes, the current reference actually includes 94 distinct genome sequences,” explains NHGRI.
The project is led by the Human Pangenome Reference Consortium, with funding from the National Institutes of Health (NIH).
The previous map of the human genome was based primarily on the DNA of one mixed-race man from Buffalo, New York, which accounted for 70% of the map, explains the New York Times. The rest of the map came primarily from people of European descent.
The updated pangenome includes both men and women “including African Americans, Caribbean Islanders, East Asians, West Africans and South Americans,” continues The Times. The update adds “nearly 120 million previously missing DNA letters” to the code.
“A richer human reference map promises to improve our understanding of genomics and our ability to predict, diagnose and treat disease. A more diverse human reference map should also help ensure that the eventual applications of genomic research and precision medicine are effective for all populations,” explained the Human Pangenome Reference Consortium in Nature on May 10.
The next steps for the pangenome
The updated map is “an exceptional advance,” evolutionary geneticist Mashaal Sohail told Smithsonian Magazine.
However: “There’s still a lot more variation that needs to be added to the pangenome to really, truly be representative of everyone,” particularly Latin Americans and Indigenous Americans, said University of Maryland geneticist Timothy O’Connor.
The map “represents a major achievement,” noted the researchers in Nature. However, “no single genome can represent the genetic diversity of our species.”
The researchers aim to include 350 people in the pangenome by mid-2024, “with a goal of reaching 700 distinct genome sequences by the completion of the project,” explained NHGRI.
“Basic researchers and clinicians who use genomics need access to a reference sequence that reflects the remarkable diversity of the human population. This will help make the reference useful for all people, thereby helping to reduce the chances of propagating health disparities,” said NHGRI Director Eric Green, M.D., Ph.D. “Creating and enhancing a human pangenome reference aligns with NHGRI’s goal of striving for global diversity in all aspects of genomics research, which is crucial to advance genomic knowledge and implement genomic medicine in an equitable way.”